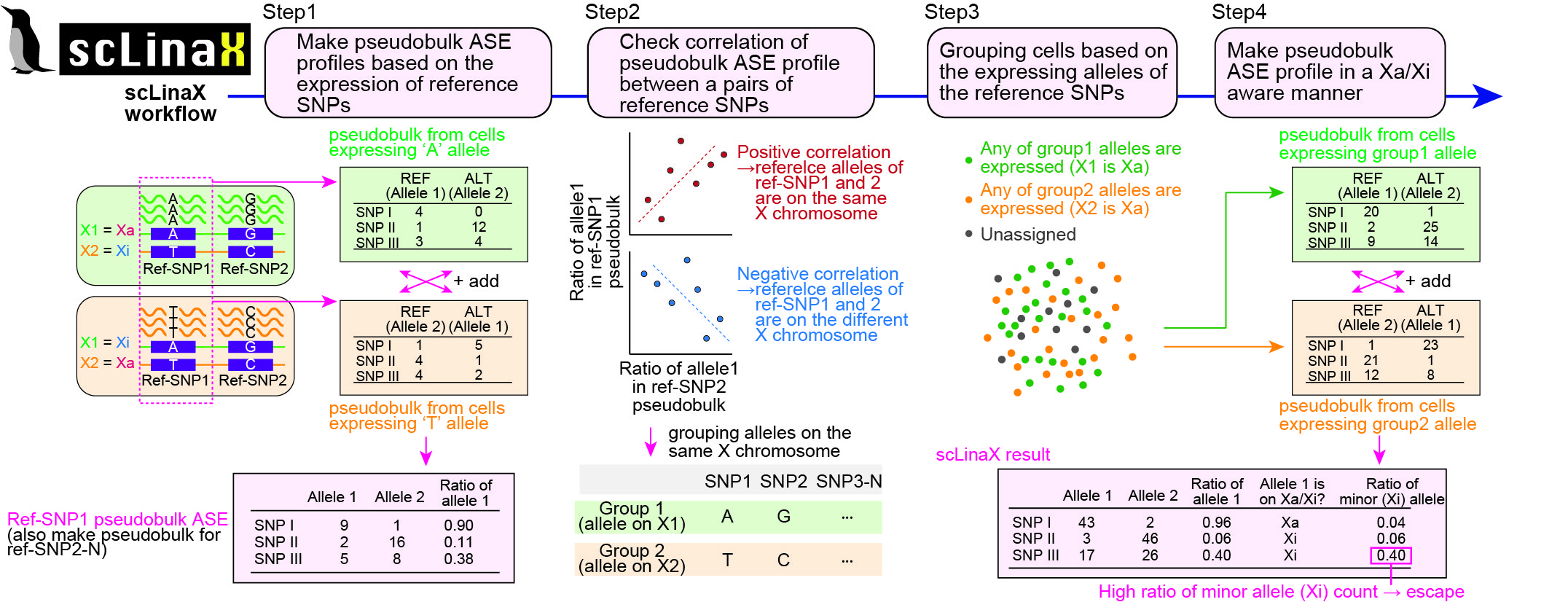
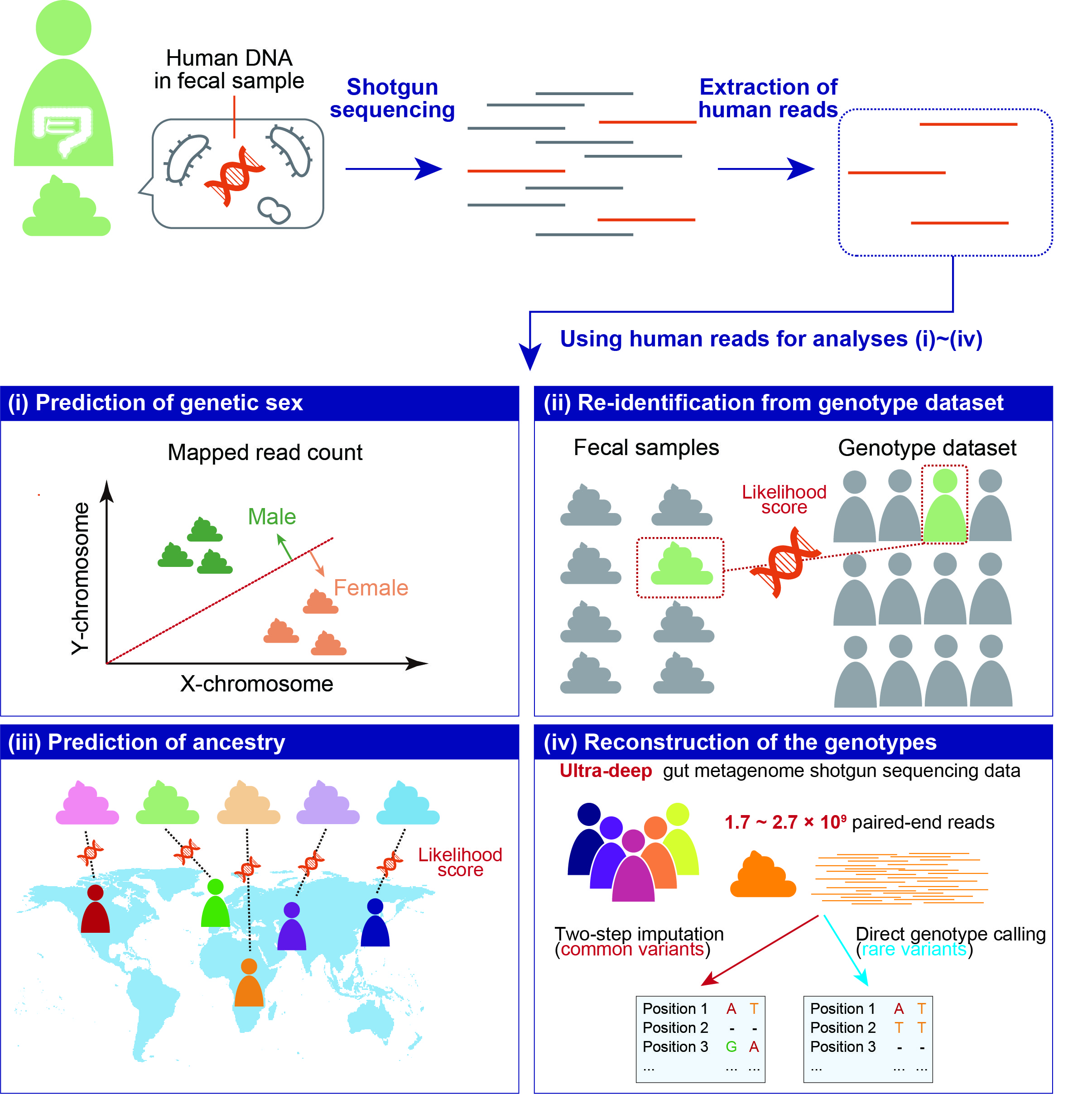
The ‘*’ and ‘†’ symbols indicate equal contribution and correspondence, respectively, in the following list.
Tomofuji, Y.†, Sonehara, K., Kishikawa, T., Maeda, Y., Ogawa, K., Kawabata, S., Nii, T., Okuno, T., Oguro-Igashira, E., Kinoshita, M., Takagaki, M., Yamamoto, K., Kurakawa, T., Yagita-Sakamaki, M., Hosokawa, A., Motooka, D., Matsumoto, Y., Matsuoka, H., Yoshimura, M., Ohshima, S., Nakamura, S., Inohara, H., Kishima, H., Mochizuki, H., Takeda, K., Kumanogoh, A. & Okada, Y.† Reconstruction of the personal information from human genome reads in gut metagenome sequencing data. Nature Microbiology 8, 1079–1094 (2023).
Edahiro, R.*, Shirai, Y.*, Takeshima, Y., Sakakibara, S., Yamaguchi, Y., Murakami, T., Morita, T., Kato, Y., Liu, Y.-C., Motooka, D., Naito, Y., Takuwa, A., Sugihara, F., Tanaka, K., Wing, J. B., Sonehara, K., Tomofuji, Y., Japan COVID-19 Task Force, Namkoong, H., Tanaka, H., Lee, H., Fukunaga, K., Hirata, H., Takeda, Y., Okazaki, D., Kumanogoh, A.† & Okada, Y.† Single-cell analyses and host genetics highlight the role of innate immune cells in COVID-19 severity. Nature Genetics 55, 753–767 (2023).
Tomofuji, Y.*, Suzuki, K.*, Kishikawa, T., Shojima, N., Hosoe, J., Inagaki, K., Matsubayashi, S., Ishihara, H., Watada, H., Ishigaki, Y., Yamanashi, Y., Furukawa, Y., Morisaki, T., Kamatani, Y., Muto, K., Nagai, A., Obara, W., Yamaji, K., Takahashi, K., Asai, S., Takahashi, Y., Suzuki, T., Sinozaki, N., Yamaguchi, H., Minami, S., Murayama, S., Yoshimori, K., Nagayama, S., Obata, D., Higashiyama, M., Masumoto, A., Koretsune, Y., Inohara, H., Murakami, Y., Matsuda, K., Okada, Y.†, Yamauchi, T.†, Kadowaki, T.†, & The BioBank Japan Project. Identification of serum metabolome signatures associated with retinal and renal complications of type 2 diabetes. Communications Medicine 3, 5 (2023).
Tomofuji, Y.†, Kishikawa, T., Maeda, Y., Ogawa, K., Otake-Kasamoto, Y., Kawabata, S., Nii, T., Okuno, T., Oguro-Igashira, E., Kinoshita, M., Takagaki, M., Oyama, N., Todo, K., Yamamoto, K., Sonehara, K., Yagita, M., Hosokawa, A., Motooka, D., Matsumoto, Y., Matsuoka, H., Yoshimura, M., Ohshima, S., Shinzaki, S., Nakamura, S., Iijima, H., Inohara, H., Kishima, H., Takehara, T., Mochizuki, H., Takeda, K., Kumanogoh, A. & Okada, Y.† Prokaryotic and viral genomes recovered from 787 Japanese gut metagenomes revealed microbial features linked to diets, populations, and diseases. Cell Genomics 2, 100219 (2022).
Itotagawa, E., Tomofuji, Y., Kato, Y., Konaka, H., Tsujimoto, K., Park, J., Nagira, D., Hirayama, T., Jo, T., Hirano, T., Morita, T., Nishide, M., Nishida, S., Shima, Y., Narazaki, M., Okada, Y., Takamatsu, H. & Kumanogoh, A. SLE stratification based on BAFF and IFN-I bioactivity for biologics and implications of BAFF produced by glomeruli in lupus nephritis. Rheumatology 62, 1988–1997 (2023).
Yan, M., Komatsu, N., Muro, R., Huynh, N. C.-N., Tomofuji, Y., Okada, Y., Suzuki, H. I., Takaba, H., Kitazawa, R., Kitazawa, S., Pluemsakunthai, W., Mitsui, Y., Satoh, T., Okamura, T., Nitta, T., Im, S.-H., Kim, C. J., Kollias, G., Tanaka, S., Okamoto, K., Tsukasaki, M. & Takayanagi, H. ETS1 governs pathological tissue-remodeling programs in disease-associated fibroblasts. Nature Immunology 23, 1330–1341 (2022).
Kishikawa, T., Tomofuji, Y., Inohara, H. & Okada, Y. OMARU: a robust and multifaceted pipeline for metagenome-wide association study. NAR Genomics and Bioinformatics 4, lqac019 (2022).
Tomofuji, Y., Kishikawa, T., Maeda, Y., Ogawa, K., Nii, T., Okuno, T., Oguro-Igashira, E., Kinoshita, M., Yamamoto, K., Sonehara, K., Yagita, M., Hosokawa, A., Motooka, D., Matsumoto, Y., Matsuoka, H., Yoshimura, M., Ohshima, S., Nakamura, S., Inohara, H., Mochizuki, H., Takeda, K., Kumanogoh, A. & Okada, Y. Whole gut virome analysis of 476 Japanese revealed a link between phage and autoimmune disease. Annals of the Rheumatic Diseases 81, 278–288 (2022).
Tomofuji, Y.*, Maeda, Y.*, Oguro-Igashira, E.*, Kishikawa, T., Yamamoto, K., Sonehara, K., Motooka, D., Matsumoto, Y., Matsuoka, H., Yoshimura, M., Yagita, M., Nii, T., Ohshima, S., Nakamura, S., Inohara, H., Takeda, K., Kumanogoh, A. & Okada, Y. Metagenome-wide association study revealed disease-specific landscape of the gut microbiome of systemic lupus erythematosus in Japanese. Annals of the Rheumatic Diseases 80, 1575–1583 (2021).
Harano, Y., Ishikawa, Y., Hattori, K., Ichinose, M., Tomofuji, Y., Okano, H., Owada, G., Kimura, Y., Nanao, T., Fujimoto, J., Nishizawa, H., Iiola, Y., Osada, J., Fujiwara, M. & Kita, Y. A case of complete atrioventricular block in secondary hemophagocytic syndrome/hemophagocytic lymphohistiocytosis recovered by plasma exchange and cytokine absorbing therapy with AN69ST continuous hemodiafiltration. Immunological Medicine 43, 171–178 (2020).
Nitta, T., Tsutsumi, M., Nitta, S., Muro, R., Suzuki, E. C., Nakano, K., Tomofuji, Y., Sawa, S., Okamura, T., Penninger, J. M. & Takayanagi, H. Fibroblasts as a source of self-antigens for central immune tolerance. Nature Immunology 21, 1172–1180 (2020).
Tomofuji, Y.*, Takaba, H.*, Suzuki, H. I., Benlaribi, R., Martinez, C. D. P., Abe, Y., Morishita, Y., Okamura, T., Taguchi, A., Kodama, T. & Takayanagi, H. Chd4 choreographs self-antigen expression for central immune tolerance. Nature Immunology 21, 892–901 (2020).
Tomofuji, Y., Ishikawa, Y., Hattori, K., Fujiwara, M. & Kita, Y. Successful treatment of refractory acute lupus haemophagocytic syndrome using rituximab: a case report. Modern Rheumatology Case Reports 4, 222–228 (2020).
Nitta, T., Kochi, Y., Muro, R., Tomofuji, Y., Okamura, T., Murata, S., Suzuki, H., Sumida, T., Yamamoto, K. & Takayanagi, H. Human thymoproteasome variations influence CD8 T cell selection. Science Immunology 2, eaan5165 (2017).
Takaba, H., Morishita, Y., Tomofuji, Y., Danks, L., Nitta, T., Komatsu, N., Kodama, T. & Takayanagi, H. Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell 163, 975–987 (2015).
The ‘*’ and ‘†’ symbols indicate equal contribution and correspondence, respectively, in the following list.